Sample Size Calculation for Case Control With Continuous Exposures
The input to program DAYSMITH
The sample sizes computed are for the detection of a given disease-exposure odds ratio, that is, the sample sizes at which a certain statistical test will reject the null hypothesis that the odds ratio is one. The input items are as follows:
R E = the odds ratio to be detected (typically a minimum value),
S = 1 or 2 for one-sided or two-sided type I error,
F = the number of controls per case,
P = the control population exposure probability, and
I = an indicator to request interaction adjustment.
Roughly speaking, interaction in statistics corresponds to effect modification in epidemiology. By not selecting an interaction adjustment, we effectively assume that the disease-exposure odds ratio does not differ across confounder levels. Interaction is discussed further below.
The number of confounder levels, denoted K is asked for next. If K = 1, unadjusted sample sizes only are computed, and no other input is required. Program DESIGN is identical to this point. For most applications, no confounder adjustment is required and so the program returns unadjusted sample sizes and is finished after a 1 is entered for K. The unadjusted formula [1] is more accurate than the usual unadjusted formulas [4, 5], and may therefore produce different sample sizes than those.
If K > 1, one of the levels of the confounder is taken to be a reference level, and is referred to as level one. The order of the levels is otherwise immaterial. The input required next is three numbers for each of the K–1 remaining levels, p 1i , p 2i , and R Ci , i = 2,..., K, which are
p 1i = Pr(C i |E) = among the exposed population, the proportion at level i of the confounder,
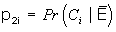
= among the unexposed population, the proportion at level i of the confounder, and
R Ci = the disease-confounder odds ratio (with confounder level i versus level 1).
For the reference level, we set R c1= 1 for the formulas that follow. We compute
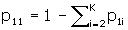
and 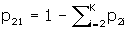 .
.
Input for program DESIGN
Whereas DAYSMITH asks for the same input as requested in the original references [1–3], we found that alternative input made more sense for our initial applications [6, 7], so a second program was written. The input for DESIGN is the same as for DAYSMITH up to the point after which the number of levels of the confounder, K, is asked for.
Again, one of the levels of the confounder is taken to be a reference level, and is referred to as level one. The input that is required next is one number for the reference level, r i , and then three (four when interaction is included) numbers for each of the K–1 remaining levels, r i , p i , and R Ci , i = 2,...,K, which are
r i = Pr(E|C i ) = the probability of exposure at level i of the confounder,
p i = Pr(C i ) = the probability of being in level i of the confounder, and
R Ci = the odds ratio of disease and confounder level i (versus level 1).
For the reference level, we again set R Ci = 1.
From Bayes Theorem, we compute
p1i=ripi/P and
p2i=(1 – ri)pi/(1 – P).
We have one more input item than is actually required, and that is used for a check, where we can use the fact that
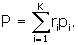
What we actually do is check the sum
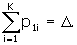
The sum Δ is supposed to be equal to one. If it is not one, then we re-define and report
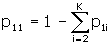
and
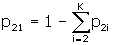
,
unless they are negative. An alternative used in earlier versions was to compute
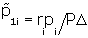
and
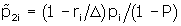
and replace
p ji
with
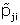
for j = 1,2 and i = 1,...,K.This is equivalent to replacing
r i
with
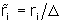
,
i = 1,..., K, which is how the program used to report the change.
An example, adjusting for a confounder
The following example is one of several computations performed for a published research protocol for a study of the association of oral contraceptive (OC) use with cardiovascular risks, controlling for age group [6]. A related protocol [7] has smoking as a confounder.
The numbers entered for P, r i , p i , and R Ci , i = 2,...,K, are all taken from the Saskatchewan government medical database, which includes the entire population from which a case-control sample is to be taken. In many applications, such numbers are not available from a reliable source. In that case, one may try sets of alternative minimum and maximum numbers for a range of results. The maximum sample sizes obtained from such sensitivity analyses would be the conservative recommendation.
Both programs first request R E to I. For R E , the outcome of interest is hospitalisation due to certain cardiovascular risks. The exposure is a specific OC with 10% of the market share [7]. Since overall OC prevalence is 30%, then P = .03 for that specific OC. Using > to denote the cursor for computer entry, we type:
>2 2 3 .03 0
for R E , S, F, P and I, respectively, then press enter. We then receive the message:
Type the number of confounder levels, and <enter>. Type 1 if no confounder.
We enter 5 levels and press enter.
>5
Now type in the population exposure probability for the reference level of the confounding variable.
This will be put at level 1, so it is Pr(E|C1)
The confounder levels are five age groups, and level 1 corresponds to the youngest age group 15–21, for which we enter the prevalence for a specific OC with 10% of the market share. We type .055 and press enter.
> .055
The reply is:
Now type in, for each of the other 4 level(s) of the confounding variable, Pr(E|Ci), Pr(Ci), and Rc(i), separated by at least one blank or <enter>, where Pr(E|Ci) = in the population at level i of the confounder, the proportion exposed, Pr(Ci) = the probability of being at level i, and Rc(i) = odds ratio of disease and confounder level i (versus level 1).
The following numbers are entered for age groups 22–26, 27–31, 22–39 and 40+:
> .038 .24 2
> .021 .2 8
> .008 .18 8
> .004 .15 28.5
Note that Rc(5) = R C5 = 28.5, a very high value. That is to be expected if all older women are included. (For the final protocol [6], a cut-off was made at age 45.) When enter is pressed, we receive some confirmation of the input, and a message that the result is written to file design.out. That is, as currently written, the sample sizes and other output are not automatically shown on the screen, but are saved in "design.out" to be viewed directly there. Appendix 2 (Second attached file, app2.txt, a text file) shows the output from the preceding session, which includes a correction of the input values.
Looking at Appendix 2, we see unadjusted sample sizes, those adjusted for age in an unmatched study, and a third set of sample sizes for a matched case-control study. For our example [6], both unmatched and matched designs are considered. With the low value of P and the high value R C5, we see that a large difference in sample sizes required for either design may result. In most applications, however, the differences are not so dramatic.
Adjusting for a matching confounder
Epidemiological literature usually gives formulas for matching which are based on the strong assumption that all sources of extraneous variation among a case and its controls are accounted for [1, 8, 9]. A third program DESIGNM was written to compute such a formula (from [1], p.294), but DESIGNM does not adjust for a confounding variable, and that strong assumption of implicit matching is rarely justified in case-control studies, so this program was not made freely available. Software which compute sample sizes for conditional logistic regression, such as EGRET SIZ[10], are alternatives to DESIGNM, which is based on Miettinen's test of the Mantel-Haenszel odds ratio for matched case-control designs. The adjustment in DAYSMITH and DESIGN is for stratified matching [2, 11, 12], where matching is by confounders. This presumes that the eventual analysis will be unconditional [2] and will account for the stratification. Consequently, it is not required that F controls be linked with each case, only that the total number of controls be F times the total number of cases.
Interaction
The literature [1, 3, 13, 15] discusses stratified analysis interaction adjustment only for confounders with K = 2. It is easy, however, to modify the formulas for multi-level interaction. Every occurrence of R E in the formulas (Appendix 1) is replaced by R E R Ij , where R Ij is the interaction factor corresponding to the j th level, j = 2,..., K. (For ∑', put R Ij inside the first sum.) We set R I1 = 1.
For two confounder levels, R I2, which is R I in Smith and Day's notation [3], is the multiplicative factor by which the odds ratio for those exposed and in level 2 of the confounder is different from the odds ratio when there is confounder-exposure-disease interaction. For R Ij , contrast is between level j and the reference level (level one).
This adjustment was made available for sensitivity analysis; specifically, to explore how much the sample size result could change if the confounder were in fact an effect modifier. Nevertheless, the adjusted formulas have been used to determine sample size in the presence of gene-environment interaction [13].
Source: https://bmcmedresmethodol.biomedcentral.com/articles/10.1186/1471-2288-1-11
0 Response to "Sample Size Calculation for Case Control With Continuous Exposures"
Post a Comment